HighPrep PCR
Paramagnetic bead-based post PCR clean-up reagent designed for efficient DNA purification and consistent DNA fragment size selection for NGS. HighPrep PCR is adaptable to most liquid handling stations and used for applications such as sequencing, NGS, microarrays, PCR, and enzymatic reactions.
Post-PCR and post-enzymatic reaction clean-up used for/during:
- NGS library preparation
- DNA size selection for NGS
- Microarrays
- PCR
- Restriction enzyme digestions, adapter ligations
- Cloning
- Rapid and reliable post-PCR and DNA clean-up reagent
- Achieve uniform and consistent DNA fragments
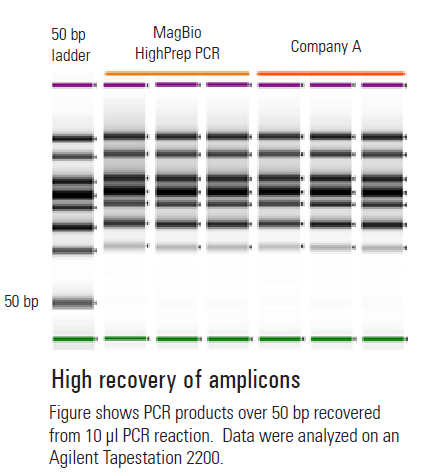
- High recovery of amplicons >100bp
- Uniform fragments size distribution
- Adaptable to high throughput liquid handling workstations
HighPrep PCR is a paramagnetic beads-based post PCR clean-up reagent designed for efficient purification of amplicons and tight size-specific selection of DNA fragments. The purification consists of removal of salts, primers, primer-dimers, dNTPs, as DNA fragments are selectively bound to the magnetic bead particles. Highly purified DNA is eluted with low salt elution buffer or water which can be used directly for downstream applications, such as library construction, next generation sequencing (NGS), Sanger sequencing, cloning, restriction digestions, adapter ligation, microarrays, and so on. This protocol can be used for manual procedures as well as in automation.
| Product information area | CE-IVD and UKCA Registered |
|---|
This is a short overview of the protocol, for all detailed protocols, please refer to PRODUCT DOCUMENTS
- Add HighPrep™ PCR reagent to the reaction mixture
- Bind DNA to paramagnetic beads
- Separation of beads + DNR from contaminants
- Wash beads with Ethanol to remove contaminants
- Elute purified PCR products or other DNA fragments from beads
| Main functions | Purifies DNA and amplicons for next generation sequencing and other applications Selectively binds fragments of 100 bp and larger depending on sample to bead ratio – performs size selection by changing the bead ratio |
| Starting material (Sample matrix) | DNA, PCR products, cDNA, gDNA |
| Starting amount | Scalable |
| RNA recovery | >70% recovery |
| Process method | Manual or automation |
| Instrument | Adaptable to most nucleic acid purification instruments but a ready-made script for Kingfisher Flex is available upon request |
| Purification method | Magnetic bead-based technology |
| RNA Purity | DNA eluted is ready for downstream processes such as real time quantitative PCR (qPCR), next generation sequencing (NGS), cloning, restriction digestions, microarrays, etc. |
| Elution Volume | 15 µl or above |
- Single-cell polyadenylation site mapping reveals 3′ isoform choice variability. Lars Velten, Simon Anders, Aleksandra Pekowska, Aino I Järvelin, Wolfgang Huber, Vicent Pelechano and Lars M Steinmetz
- RNA-seq Analysis of Irrigated vs. Water Stressed Transcriptomes of Zea mays Cultivar Z59. Divya Bhanu, Kandasamy Ulaganathan, Arun K. Shanker and S. Desai
- Nucleotide pools dictate the identity and frequency of ribonucleotide incorporation in mitochondrial DNA. Anna-Karin Bergl, Clara Navarrete, Martin K. M. Engqvist, Emily Hoberg, Zsolt Szilagyi, Robert W. Taylor, Claes, M. Gustafsson, Maria Falkenberg, Anders R. Clausen
- CleanTag small RNA library kit with no Gel Purification. Mark Livingstone
- Real-time, portable genome sequencing for Ebola surveillance. Joshua Quick
- RNA-Seq analysis of urea nutrition responsive transcriptome of Oryza sativa elite indica cultivar RP Bio 226. Mettu Madhavi Reddy, Kandasamy Ulaganathan
- Taxonomic description and genome sequence of Bacillus campisalis sp. nov., a member of the genus Bacillus isolated from a solar saltern. Rajendran Mathan Kumar,Gurwinder Kaur, Anand Kumar, Monu Bala, Nitin Kumar Singh, Navjot Kaur, Narender Kumar and Shanmugam Mayilraj
- Genome Mining and Comparative Genomic Analysis of Five Coagulase- Negative Staphylococci (CNS) Isolated from Human Colon and Gall Bladder. Ramesan Girish Nair, Gurwinder Kaur, Indu Khatri, Nitin Kumar Singh, Sudeep Kumar Maurya, Srikrishna Subramanian, Arunanshu Behera, Divya Dahiya, Javed N Agrewala and Shanmugam Mayilraj
- Draft genome sequence of Oryza sativa elite indica cultivar RP Bio-226. Mettu M. Reddy and Kandasamy Ulaganathan
- Having older siblings is associated with gut microbiota development during early childhood. Martin Frederik Laursen, Gitte Zachariassen, Martin Iain Bahl, Anders Bergström, Arne Høst, Kim F. Michaelsen and Tine Rask Licht
- Infant Gut Microbiota Development Is Driven by Transition to Family Foods Independent of Maternal Obesity. Martin Frederik Laursen, Louise B. B. Andersen, Kim F. Michaelsen, Christian Mølgaard, Ellen Trolle, Martin Iain Bahl, Tine Rask Licht.
- Unbiased Identification of Blood-based Biomarkers for Pulmonary Tuberculosis by Modeling and Mining Molecular Interaction Networks. Awanti Sambarey, Abhinandan Devaprasad, Abhilash Mohan, Asma Ahmed, Soumya Nayak, Soumya Swaminathan, George D’Souza, Anto Jesuraj, Chirag Dhar, Subash Babu, Annapurna Vykarnam, Nagasuma Chandra
- Knock-In of a 25-Kilobase Pair BAC-Derived Donor Molecule 9 by Traditional and CRISPR/Cas9-Stimulated Homologous Recombination. Tiffany Leidy-Davis, Kai Cheng, Leslie O. Goodwin, Judith L. Morgan, Wen Chun Juan, Xavier Roca, Sin-Tiong Ong, David E. Bergstrom
- Phosphopantothenoylcysteine Synthetase from Escherichia coli. Erick Strauss, Cynthia Kinsland, Ying Ge, Fred W. McLafferty, and Tadhg P. Begley
- Microbial diversity and community respiration in freshwater sediments influenced by artificial light at night. Franz Hölker, Christian Wurzbacher, Carsten Weißenborn, Michael T. Monaghan, Stephanie I. J. Holzhauer, Katrin Premke
- Novel Molecular Findings in Protein Tyrosine Phosphatase Receptor Gamma (PTPRG) Among Chronic Myelocytic Leukemia (CML) Patients Studied By Next Generation Sequencing (NGS): A Pilot Study in Patients from the State of Qatar and Italy. Maria Monne, Mohammed Araby, Ali Sayab, Marzia Vezzalini, Luisa Tomasello, Helmout Modjtahedi.
- Molecular and Biochemical Characterization of AtPAP15, a Purple Acid Phosphatase with Phytase Activity, in Arabidopsis. Ruibin Kuang, Kam-Ho Chan, Edward Yeung and Boon Leong Lim
- Effect of humic acid on anaerobic digestion of cellulose and xylan in completely stirred tank reactors: inhibitory effect, mitigation of the inhibition and the dynamics of the microbial communities. Samet Azman & Ahmad F. Khadem & Caroline M. Plugge & Alfons J. M. Stams
- Bacterial Community Profiling of Plastic Litter in the Belgian Part of the North Sea Caroline. De Tender, Lisa I. Devriese, Annelies Haegeman, Sara Maes, Tom Ruttink, and Peter Dawyndt
- Taxonomic description and genome sequence of Salinicoccus sediminis sp. nov., a halotolerant bacterium isolated from marine sediment. Rajendran Mathan Kumar, Gurwinder Kaur, Narender Kumar, Anand Kumar, Nitin Kumar Singh, Monu Bala, Navjot Kaur, Shanmugam Mayilraj
- The role of the intestinal microbiota in pneumonia and sepsis. Lankelma, J.M.
- Taxonomic description and genome sequence of Salinicoccus sediminis sp. nov., a halotolerant bacterium isolated from marine sediment. Rajendran Mathan Kumar, Gurwinder Kaur, Narender Kumar, Anand Kumar, Nitin Kumar Singh, Monu Bala, Navjot Kaur and Shanmugam Mayilraj.
- Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Anders R Clausen, Scott A Lujan, Adam B Burkholder, Clinton D Orebaugh, Jessica S Williams, Maryam F Clausen, Ewa P Malc, Piotr A Mieczkowski, David C Fargo, Duncan J Smith & Thomas A Kunkel
- Metagenomic analysis of microbial community of an Amazonian geothermal spring in Peru. Sujay Paul, Yolanda Cortez, Nadia Vera, Gretty K. Villena, Marcel Gutiérrez-Correa
- High throughput de novo RNA sequencing elucidates novel responses in Penicillium chrysogenum. Yesupatham Sathishkumar, Chandran Krishnaraj, Kalyanaraman Rajagopa, Dwaipayan Sen and Yang Soo Lee
- Growth and activity of ANME clades with different sulfate and sulfide concentrations in the presence of methane. Peer H. A. Timmers, H. C. A. Widjaja-Greefkes, Javier Ramiro-Garcia, Caroline M. Plugge and Alfons J. M. Stams
- Tryptophan restriction arrests B cell development and enhances microbial diversity in WT and prematurely aging Ercc12/D7 mice. Adriaan A. van Beek, Floor Hugenholtz, Ben Meijer, Bruno Sovran, Olaf Perdijk, Wilbert P. Vermeij, Renata M. C. Brandt, Sander Barnhoorn, Jan H. J. Hoeijmakers, Paul de Vos, Pieter J. M. Leenen, Rudi W. Hendriks and Huub F. J. Savelkoul.
- Infant Gut Microbiota Development Is Driven by Transition to Family Foods Independent of Maternal Obesity. Martin Frederik Laursen, Louise B. B. Andersen, Kim F. Michaelsen, Christian Mølgaard, Ellen Trolle, Martin Iain Bahl, Tine Rask Licht
- Supraglacial bacterial community structures vary across the Greenland ice sheet. Karen A. Cameron, Marek Stibal, Jakub D. Zarsky, Erkin Gozdereliler, Morten Schostag and Carsten S. Jacobsen
- ERG signaling in prostate cancer is driven through PRMT5- dependent methylation of the Androgen Receptor. Mounir, Z., J. M. Korn, T. Westerling, F. Lin, C. A. Kirby, M. Schirle, G. McAllister
- 2 1-fructans modulate the immune system in vivo by direct interaction with the mucosa in a microbiota-independent fashion. Fransen, Floris; Sahasrabudhe, Neha; Elderman, Marlies; Bosveld, Margaret; El Aidy, Sahar Farouk Abdelsalam; Hugenholtz, Floor; Borghuis, Theo; Kousemaker, Ben; Winkel, Simon; van der Gaast-de Jongh
- Benzene degradation in a denitrifying biofilm reactor: activity and microbial community composition. Marcelle J. van der Waals, Siavash Atashgahi, Ulisses Nunes da Rocha, Bas M. van der Zaan, Hauke Smidt, Jan Gerritse
- Taxonomic description and genome sequence of Salinicoccus sediminis sp. nov., a halotolerant bacterium isolated from marine sediment. Rajendran Mathan Kumar, Gurwinder Kaur, Narender Kumar, Anand Kumar, Nitin Kumar Singh, Monu Bala, Navjot Kaur, Shanmugam Mayilraj
Want to share one more referenece? Write us an email.



 (301) 302-0144
(301) 302-0144 info@magbiogenomics.com
info@magbiogenomics.com