4 DNA Quantification Methods to Consider
Your DNA prep is done and you’re for the next step in your experiment. But first, you need to check for the presence of DNA in your tube even if you can’t see it yet! There are many ways to do this and researchers select the method based on the downstream application and time availability.
UV absorbance
UV absorbance is a common DNA quantification method. It involves measuring the absorbance or transmission of light through a liquid to calculate the concentration of substances in that liquid. Different molecules absorb different wavelengths of light in varying degrees. The spectrophotometer is used to measure these absorbances. First the absorbance of the buffer the DNA is in is measured. Then the absorbance of the DNA sample is measured.
These absorbances present an idea of the DNA prep concentration as well as contaminants if any. Nucleic acids absorb maximally at 260 nm; proteins at 280 nm and organic compounds and chaotropic salts are maximally absorbed in 230 nm. The A260/A280 ratio should be between 1.8 and 2.0 while the A260/A230 ratio should be more than 1.5.
Then, the DNA concentration can be estimated by multiplying the A260 reading with the dilution factor.
This method is simple, fast and doesn’t require any special reagents. However, at low concentrations of DNA, it can’t distinguish between DNA and RNA.
Fluorescence dyes
Dyes that fluoresce when they bind to DNA can also be used to quantify DNA. This method is more sensitive than UV absorbance, especially at low DNA concentrations. It can also be used to quantify DNA for NGS (next-generation sequencing).
Fluorescence-based methods require a standard curve to compare the fluorescence of your sample against known values, and hence, quantify your DNA prep. Some fluorometers can also calculate your sample concentration.
Agarose gel electrophoresis
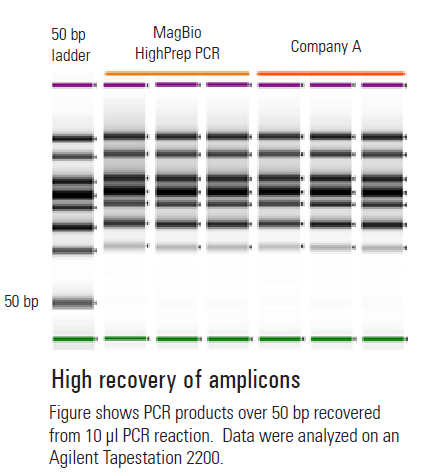
This is a slower method to quantify DNA but it can help you quantify DNA as well as determine whether the DNA is intact and the right size. Absorbance-based methods can’t do this.
The method involves pouring your gel containing a DNA intercalating dye and selecting a DNA ladder with known concentrations. Samples of DNA are run at different dilutions and quantified based on band intensities relative to the intensity in the ladder. This method is best suited for DNA fragments (as in PCR).
Capillary electrophoresis
Capillary electrophoresis-based DNA quantification is similar to the gel electrophoresis method but it is automated.
During the run, DNA fragments move through a micro-fluidic channel and the sample is separated by electrophoresis. This method is often used before NGS or microarray studies rather than the standard plasmid prep.
You need to consider several factors, such as cost, time, equipment, and expected DNA concentration, while selecting your DNA quantification method. Compare the benefits and disadvantages of each method for your application to check which method might be more suited than another.



 (301) 302-0144
(301) 302-0144 info@magbiogenomics.com
info@magbiogenomics.com