HighPrep DTR
Magnetic beads based reagent for manual and automated Sanger sequencing reaction clean-up for both ABI and MegaBACE sequencing platforms.
Sanger sequencing reaction clean-up / dye terminator removal for:
- Sanger sequencing on ABI platforms
- Sanger sequencing on MegaBACE platforms
- Rapid and reliable clean-up of sequencing reactions
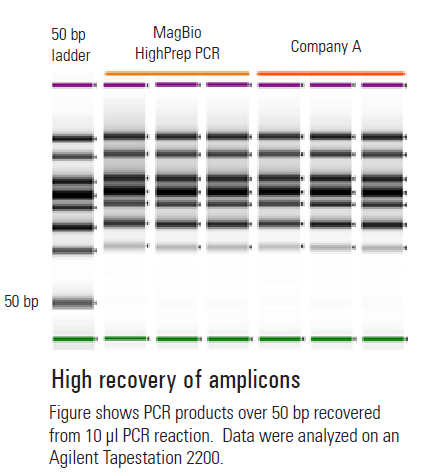
- High DNA recovery yields, long sequence readings
- Flexible - can be used in any format
- No centrifugation step, no filtration step
- Adaptable to common liquid handling workstations
The HighPrep DTR is a high performance paramagnetic bead-based reagent for BigDye™ terminator removal from Sanger sequencing reactions for reliable and long readings of DNA sequences. The HighPrep DTR consists of a selective binding of DNA to the paramagnetic beads, followed with washing off nucleotides, primers, and non-targeted amplicons, and finally elution of pure DNA. HighPrep DTR is an excellent choice for both manual and fully automated purification of sequencing reaction products. The protocol can be adapted to common liquid handling workstation.
BigDye is a registered trademark of Applied Biosystems
This is a short overview of the protocol, for all detailed protocols, please refer to PRODUCT DOCUMENTS
- Add HighPrep™DTR reagent to sample
- Add freshly prepared 85% ethanol
- Bind sample to paramagnetic beads
- Wash beads with 85% ethanol to remove contaminants
- Elute purified DNA from beads
| Features | Specifications |
| Main functions | Removes unincorporated dye terminators from Sanger sequencing reactions |
| Starting material (Sample matrix) | DNA, PCR products, cDNA, gDNA |
| Starting amount | Scalable |
| Process method | Manual or automation |
| Purification method | Magnetic bead-based technology |
| Application | Sanger sequencing on ABI platforms and MegaBACE platforms |
| Elution Volume | 15 uL and above |
- An economical and efficient method of post reaction cleanup of labeled dye terminator sequencing products. Rajesh R Kundapur, Vijay Nema
- Knock-In of a 25-Kilobase Pair BAC-Derived Donor Molecule 9 by Traditional and CRISPR/Cas9-Stimulated Homologous Recombination. Tiffany Leidy-Davis, Kai Cheng, Leslie O. Goodwin, Judith L. Morgan, Wen Chun Juan, Xavier Roca, Sin-Tiong Ong, David E. Bergstrom
- Nuclear introgression without mitochondrial introgression in two turtle species exhibiting sex-specific trophic differentiation. Sarah M. Mitchell,nLaura K. Muehlbauer, Steven Freedberg
- A European epidemiological survey of Vibrio splendidus clade shows unexplored diversity and massive exchange of virulence factors. Nasfi H, Travers MA, de Lorgeril J, Habib C, Sannie T, Sorieul L, Gerard J, Avarre JC, Haffner P, Tourbiez D, Renault T, Furones D, Roque A, Pruzzo C, Cheslett D, Gdoura R, Vallaeys T.
- Factors affecting Sanger sequencing robustness and data quality. Patrick Warner, Shea Anderson, Archana Deshpande, Daryl M. Gohl, Kenneth B. Beckman
Want to share one more referenece? Write us an email.



 (301) 302-0144
(301) 302-0144 info@magbiogenomics.com
info@magbiogenomics.com